February 27, 2025
Co-authors are Education and Theory Working Group Participants, Resource Developers, and Testers of the Review Materials:
Jody Peters1, Abby Lewis2, Alyssa Willson1, Cazimir Kowalski1, Cole Brookson3, Gerbrand Koren4, Hassan Moustahfid5, Hannah O’Grady1, Jason McLachlan1, John Zobitz6, Mary Lofton7, Ruby Krasnow8, Saeed Shafiei Sabet9
1University of Notre Dame, 2Smithsonian Environmental Research Center, 3Yale University, 4Utrecht University, 5NOAA, 6Augsburg University, 7Virginia Tech, 8University of Maine, 9University of Guilan
The goal of this blog post is to share resources that individuals in the EFI community have developed and have found useful when reviewing code.
Specifically, this blog post provides
- An Overview of why to review code or have your code reviewed
- The Background for this blog post and the resources presented
- Resources developed and tested by blog co-authors including a project overview template and code review checklist template
- Pain points to be aware of and suggestions for how to manage them in the review process
- Other resources from SORTEE (Society for Open, Reliable, and Transparent Ecology and Evolutionary Biology)
- Additional resources the EFI working groups have found useful
- Best wishes for your code review
Why to review code or have your code reviewed
Just like text review, such as peer-review or co-author review, improves published manuscripts, code review can be critical for reliability, reusability, reproducibility, and knowledge sharing. Code review can take many forms, including as an individual or team activity, in research or classroom settings. Ultimately, reviewing code provides an opportunity to:
- learn from more experienced coders about how to code or code more efficiently, either as the code reviewer or as the one requesting code review
- provide another set of eyes to reduce errors and the potential of reporting faulty results, which can slow down scientific progress and may lead to retractions for a publication
- increase the reliability and reusability of the code to help with the repeatability of studies and the application of previously developed code in new contexts. This is increasingly recognized as an important characteristic of research software (Barker et al., 2022)
- carefully check any code that has been drafted with AI tools (e.g., chatGPT, Copilot, etc.). AI tools may be helpful to save time when first drafting code. However, any code created using an AI tool should not be blindly trusted to work. Ben Weinstein discusses this at time 13:49 in the January 2025 Statistical Methods Seminar Series presentation on the DeepForest package https://youtu.be/fhlC0W2kDMQ?si=KZYObPIlt2512T1Y
Open code reviews coordinated through a third party like rOpenSci also provide opportunities to network and meet colleagues and collaborators from other scientific domains. For R package developers, submitting your code to rOpenSci for peer review has many additional benefits, including assistance with package maintenance and social media promotion.
While there are many benefits of having your code reviewed, there are, however, few resources and standards that exist for code review in ecology, and the specific methods for code review will likely differ across career stages, manuscript development stages, etc.
Background
Over the past year, the EFI Theory and Education working groups have discussed and developed resources for reviewing code that we wanted to share with others who are thinking about or are in the process of having their code reviewed or reviewing code for others.
The working group discussions and subsequent resources were framed around the Ivimey-Cook et al 2023 paper “Implementing code review in the scientific workflow: Insights from ecology and evolutionary biology” (https://doi.org/10.1111/jeb.14230) and materials shared by the SORTEE community (Society for Open, Reliable, and Transparent Ecology and Evolutionary Biology; https://www.sortee.org/).
The Ivimey-Cook et al 2023 paper provides commentary on
- How to effectively review code
- How to set up projects to enable this form of review
- How to implement code review at several stages throughout the research process
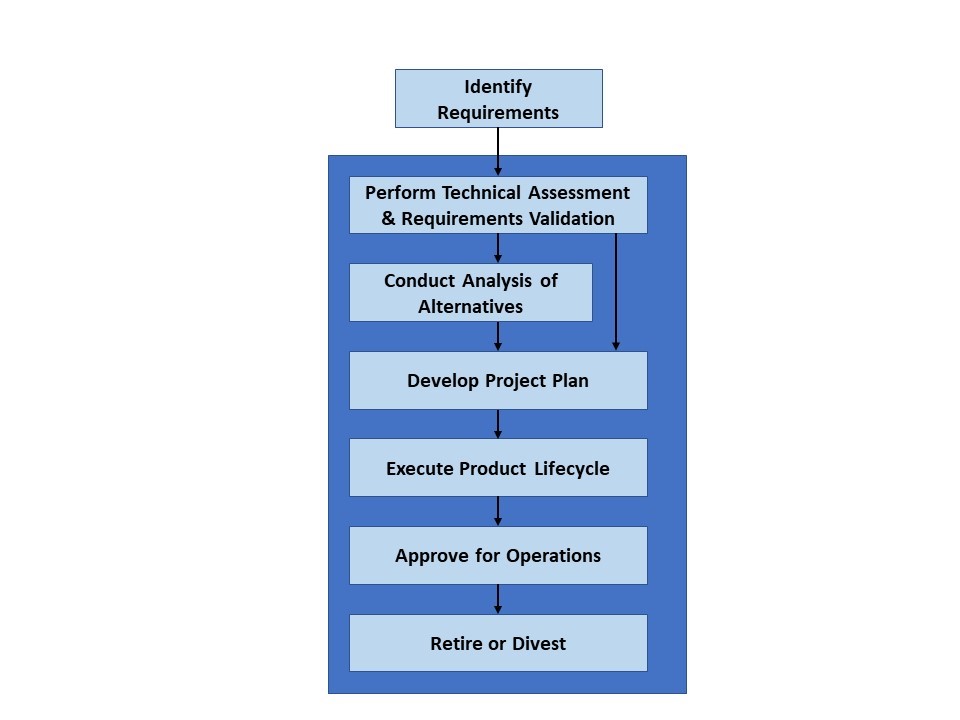
In the paper, the authors highlight “The 4 Rs” that code review should evaluate:
- Is the code as Reported?
- Methods and code must match
- Does the code Run?
- Code must be executable
- Is the code Reliable?
- Code runs and completes as intended
- Are the results Reproducible?
- Results must be able to be reproduced
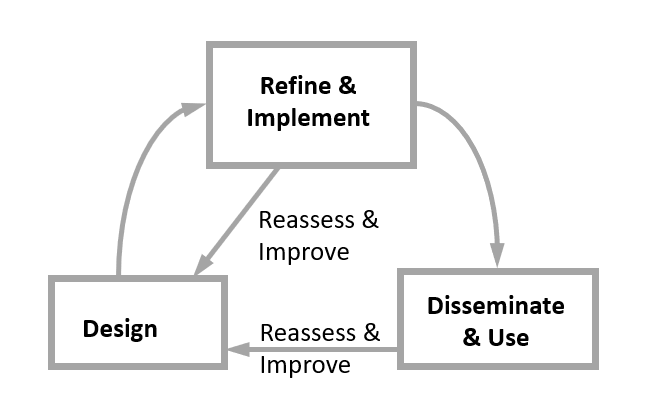
They describe a basic workflow of questions to answer when reviewing code, as summarized in the figure below.
(Image source: Ivimey-Cook et al., 2023)
Jump to the top of the blog post
Resources developed by working group members to share with the EFI community
Based upon the work by Ivimey-Cook et al., the EFI Education and Theory working groups put together two documents: a project overview document and a code review checklist. During the creation of these documents, the assumption is that the code review is being done by an internal code reviewer (e.g., lab mate) vs an external code reviewer (e.g., reviewer for a journal).
The project overview template is filled out by the person who wrote the code. This document helps clarify the purpose of the analysis and where feedback would be useful.
Conversely, the checklist template is filled out by the person who is reviewing the code. It identifies the key points to check during the review.
- Project Overview to Prep for Code Review Template – This Project Overview template helps authors describe their project for individuals who will be reviewing their code.
- Code Review Checklist Template (spreadsheet version, pdf version) – This Checklist template is based on the material in Ivemy-Cooke et al 2023. There are checklists related to project organization, project and input metadata, code readability, and output readability that both authors and reviewers can check and add notes about. This Checklist has been implemented by working group members based at the University of Notre Dame.
While the checklist is a good way for a reviewer to check off what has been reviewed, we recommend creating a separate document to note any issues a reviewer has during the code review that needs to be addressed by the code author. The code review document can be a word or Google doc or it can be an RMarkdown (.Rmd) file in the GitHub repo with the code. The benefit of the .Rmd file versus a pull request is that the updates to the .Rmd file allow for versioning and transparency without requiring the code reviewer to make the actual code fixes, but instead leaving that to the code author.
Jump to the top of the blog post
Pain points to be aware of & suggestions for how to manage them
Pain point 1: Code that takes a long time to run or creates a large amount of output
How to Manage: Code authors can provide aggregated output or a small example data set that can be run locally
Often in ecological forecasting we develop code workflows that take hours, days, or weeks to run. To avoid placing this computational burden on a code reviewer, authors can either provide the aggregated analysis output or a small example data set for review. Choosing between these two options likely depends on the goals of the code review. If the review is happening at a more mature stage of the project and the primary goal is to reproduce manuscript figures, providing the reviewer with aggregated output may suffice. The disadvantage of this approach is that the reviewer will likely not be running all the steps in the analysis, and therefore may miss errors that occur “upstream” of the creation of the aggregated output. On the other hand, if feedback is needed on the scientific merit and correctness of the analysis from start to finish, it may be better to provide a small example data set to allow the reviewer to run the entire workflow. The disadvantage of this approach is that the results obtained with the example data set will not match the results reported in the final manuscript.
If authors choose to provide an example subset of data for code review, tools such as RMarkdown, Quarto, or Jupyter Notebooks can be useful to walk reviewers through the analysis. These file types allow text interspersed with code and visualizations in an interactive format, which may help a reviewer navigate the steps of a complex coding workflow. The downside is that the file paths/etc will need to be updated to apply to the subset of data.
Each project will need to decide what specific approach works best for them.
Pain point 2: Large data used in analyses that are not yet publicly archived. It can be logistically challenging to share the data and it makes checking paths, folder structure, or data intermediates difficult.
How to Manage: One approach taken by some blog co-authors is to use the staging environment in the Environmental Data Initiative data portal to make data available online so it can be sourced as a script before it is assigned a DOI. The benefit of this approach is that the data is then ready to be archived and ready to add to the manuscript once the checks on the code are finalized.
Another approach used is to share the data with the code reviewer using an external hard-drive or Google drive with zipped folders. If this approach is taken, be sure to include notes for where file paths need to be changed for and after the review.
Pain point 3: Different versions of R (or other coding language) or data packages and their dependencies and compiled languages, e.g. C++
How to Manage: Use a docker environment.
If using a container, code authors should be sure to provide clear instructions to peer reviewers about how to set up and run a container on their machine, as well as how to delete/uninstall the container software afterward. Because using a container adds an extra step to the review process (particularly for those who have not previously used containers), it may be best to reserve this option for analyses with a high number of software and package dependencies, because installing a container becomes easier than installing all those dependencies separately.
Pain point 4: Reviewing code can take a substantial amount of time, anywhere from a couple of hours to a couple of days, depending on the scope of the review. Blog co-authors have found that completing a code review for a co-author often takes longer than completing a peer review of the manuscript.
How to Manage: Be cognizant that reviewing code can take a substantial amount of time and plan accordingly to give the reviewer enough time before major milestones, such as submitting a manuscript. Alternatively, depending on the situation, it may be best to plan for continuous code review as part of the manuscript writing process.
No matter the approach, we highly recommend including code reviewers as co-authors for publications using output from code that was reviewed. This is to appropriately recognize the effort and intellectual contribution involved from code review and it is in line with increasing recognition for co-authorship and different roles in the development and testing process (Leem et al. 2023)
Pain point 5: Getting too much or too little input from a reviewer based on your publication needs.
How to Manage: Before you ask for a review, determine how in-depth you want the review to be. This may reflect what stage you are in the manuscript writing or analysis process. If you are early in the process and want help with making your code more efficient, the code review feedback may work best to come in as GitHub pull requests. If you feel your code is finalized and ready for one final review before publication, then it may work best to have a more in-depth review to confirm the output for the publication can be recreated with the code that will be shared for the publication. The “Project Overview” template (described above) is intended to help communicate these needs when asking for code review.
In practice, you may wish to seek code review at multiple stages in the analysis and writing process. For example, you might ask for a co-author to review key components of the analysis for code correctness (e.g., the “science” is correct; units are converted properly; statistical analyses are applied appropriately; and so on) as you explore your preliminary results. Later, while developing and preparing to submit your manuscript, you may ask a co-author to review more surficial aspects of the code base (e.g., files are organized in a logical way; all filepaths are relative; and so on). We re-emphasize that even senior scientists and experienced coders make mistakes! It is always better to find them before publication than afterward.
Jump to the top of the blog post
Other resources from SORTEE
SORTEE is the Society for Open, Reliable, and Transparent Ecology and Evolutionary Biology. The SORTEE community led the Ivimey-Cook et al 2023 paper and in addition to the paper have shared other resources.
- SORTEE: https://www.sortee.org/
- SORTEE Slack channel – here is the link to join https://join.slack.com/t/sortee/shared_invite/zt-2fnqytett-AND1mTuXBKQWYyWUXKn6YA
- Library of code mistakes: https://docs.google.com/presentation/d/12QN3WUc5v1Df7OArEox2U7l_N_qnHHuwzjCYiI4idC8/edit#slide=id.p
- Issues that people have found when their code has been reviewed can be anonymously added to this file. It is structured with the same headings used in the 4R paper on code review
Other papers or resources the EFI working groups found helpful
- Iveimey-Cook et al. 2023. Implementing code review in the scientific workflow: Insights from ecology and evolutionary biology. Journal of Evolutionary Biology 36: 1347-1356. https://doi.org/10.1111/jeb.14230
- Filazzola, A. and C.J. Lortie. 2022. A call for clean code to effectively communicate science. Methods in Ecology and Evolution 13, 2119–2128. https://doi.org/10.1111/2041-210X.13961
- Hunter-Zinck, H. et al. 2021. Ten simple rules on writing clean and Reliable Open-source scientific software. PLOS Computational Biology 17(11): e1009481.
https://doi.org/10.1371/journal.pcbi.1009481 - Alston, J.M. and J.A. Rick. 2021. A Beginner’s Guide to Conducting Reproducible Research. Bulletin of the Ecological Society of America 102(2): 1-14. https://www.jstor.org/stable/27000718
- Git + GitHub As A Platform For Reproducible Research. GitHub – gchure/reproducible_research: A template repository for how I structure my scientific research. This repository sets out the skeleton of an organizational structure used for scientific research.
- Cooper, N. 2018. A Guide to Reproducible Code in Ecology and Evolution. https://nhm.openrepository.com/handle/10141/622618. A Guide to Reproducible Code covers all the basic tools and information you will need to start making your code more reproducible.
- Lakens, D. 2022. Improving Your Statistical Inferences. Retrieved from https://lakens.github.io/statistical_inferences/. https://doi.org/10.5281/zenodo.6409077.
An open educational resource contains information to improve statistical inferences, design better experiments, and report scientific research more transparently. - Boettiger, C. 2015. An introduction to Docker for reproducible research. ACM SIGOPS Operating Systems Review, 49, 71–79. https://doi.org/10.1145/2723872.2723882
- Reproducibility Demo: https://github.com/rqthomas/reproducibility-demo
- Barker, M. et al. 2022. Introducing the FAIR Principles for research software. Scientific Data 9: 622. https://doi.org/10.1038/s41597-022-01710-x
- Leem, D. et al. 2023. SORTÆD: Software Role Taxonomy and Authorship Definition (0.1). Zenodo. doi:10.5281/zenodo.7896456; https://sdruskat.net/software-authorship/.
Jump to the top of the blog post
Best wishes for your code review
We wish you all the best as you create and have your code reviewed or review code for others.
This XKCD comic was shared in one of the EFI working group calls and hopefully, it brings you a smile and it inspires you to avoid this in your own code. Or hopefully, you won’t see it in the code you are reviewing! (https://xkcd.com/1833/)!